Code for the ANOM and Nemenyi tests for MatLab. Download here.
For a discussion of the two tests and the various ways to visualise the results look at this post.
Here are some examples, using the M3 results:
>> anom(X,0.05,labels);

The models in red are significantly better than the average (solid line).
For the Nemenyi test:
>> nemenyi(X,1,'labels',labels,'colormap','hsv');

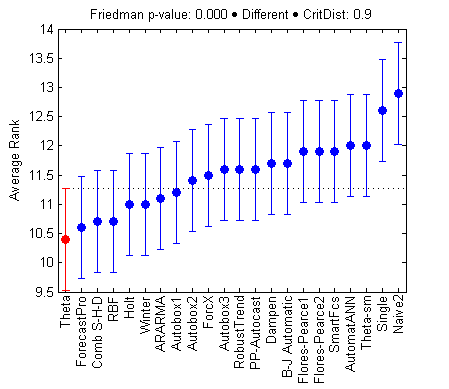
There is no evidence of significant differences for models joined by the vertical lines. Depending on the model we are considering, different groups can be formed. The groups are identified using the mean rank of a model ± the critical distance.
There are several plotting options coded for the the nemenyi.m. I have also put an option for changing the colourmap to better visualise the comparisons. A potentially useful option is to set:
>> nemenyi(X,1,'labels',labels,'colormap','omcb');
which gives you the MCB test with ordered models. These two tests use exactly the same statistic, although MCB compares only with the best model.
You may also be interested in this very nice visualisation of Nemenyi post-hoc test by Farshid Sepehrband available here.
Hi Nikolas,
I am going to use your MATLAB function “nemenyi.m”. I have called the function as below:
results = [0.01 0.02 0.03 0.04; 0.01 0.03 0.02 0.04; 0.02 0.02 0.03 0.04; 0.01 0.03 0.03 0.04; 0.01 0.02 0.02 0.03; 0.01 0.04 0.02 0.04];
labels = {‘rTree’,’SVR’,’RBF’,’FFNN’};
[p, nemenyi, meanrank, CDa, rankmean] = nemenyi(results,6,’ploton’,’omcb’)
or
[p, nemenyi, meanrank, CDa, rankmean] = nemenyi(results,6,’labels’,labels)
but I got this error for both cases:
Undefined function or variable ‘statgetargs’.
The ‘ststgetargs’ function is in line 60 inside ‘nemenyi’ function.
It should be said that when I call the function like this it works properly:
[p, nemenyi, meanrank, CDa, rankmean] = nemenyi(results,6 )
I would appreciate it if you can help me to figure it out.
Thanks,
Rahman
Hi Rahman,
I get no error, what version of MatLab are you using?
Thanks,
Nikos
Thank you for the reply Nikos. I am using 2016a. If ‘statgetarg’ is a function, there is no *.m file for this function in your toolbox.
Would you be able to send me the functions that you have used to execute those commands without getting errors?
My email address is: a.peimankar@gmail.com
Thanks
Rahman
Hi Nikos, hi Rahman,
I am also using Matlab R2016a and I also receive this error message:
Undefined function or variable ‘statgetargs’.
Error in nemenyi (line 60)
[~, ~, cmap, alph, labels, ploton] = statgetargs(okargs,defaults,varargin{:});
Did you solve the problem by sending additional files? If yes, could you please send me the necessary functions? My email address is: kue@biba.uni-bremen.de
Thank you and best regards,
Mirko
statgetargs is a build-in function, but I think at some point this internal function got superseded by other ones. This post should be helpful:
http://uk.mathworks.com/matlabcentral/newsreader/view_thread/100926
Thank you Nikos.
Hi,
As you have mentioned that “There is no evidence of significant differences for models joined by the vertical lines”. However, in the second figure, various lines are connected whose difference is more than the critical value then, how they are with the insignificant difference?
It depends on the model you are starting the comparison from, so different groups of insignificantly different models will be formed. For instance take Theta. 10.4 + 0.9 (critical distance) = 11.3, which is just enough to include Autobox1. If one the other hand I start from Autobox1 I get a different group. It starts from Theta, but reaches up until 11.2 + 0.9 = 12.1, which includes SmartFcs. Hope this helps!